Last semester I took a class in C and C++. In this class, I created an ASCII based dungeon and an adventurer who fights monsters and has to defeat a boss to win the game.
In my final semester in the Peterson Lab, I worked with Illumnia read data to attempt to assemble the B54 maize genome idependently from the B73 reference genome. The proved to a great challenge since the repetitiveness of the maize genome demanded a reference genome but we were trying to assemble areas unique to B54. After the semester, I had a draft assembly that is currently being tested in the lab.
In the Spring of 2017, I worked in a genetics research lab at Iowa State. At this time, I had just started learning Java formally in class, only really knowing basic data. However, my research demanded that I learn more which resulted in learning a lot from the internet. Looking back at my code from a more experienced standpoint, I recognize that I could have made better use of different data structures, following best practices for Java, and overall making less repetitive and more solid code.
My project in Dr. Peterson's lab involved primer analysis. Some of my work was in a typical molecular lab, extracting DNA from maize samples and testing primers in PCR reaction. However, there is a major problem when working with the maize genome. The DNA is highly repetitive and primers, short sequences of twenty DNA basepairs needed for replication, require unique sequences. Looking at each piece of DNA to see if it is a valid primer sequence is time consuming and potential locations are over looked.My process incorporated existing bioinformatics tools and java code that I wrote in order produce a map that marked regions of high similarity in the genome and low similarity in the genome, allowing my lab members to find primers faster.
I also presented my research to a team of faculty and graduate students; Take a look at my presentation. Below are graphics from my presentation. This first explains the path of my program and the second shows the final, usable results of the program.
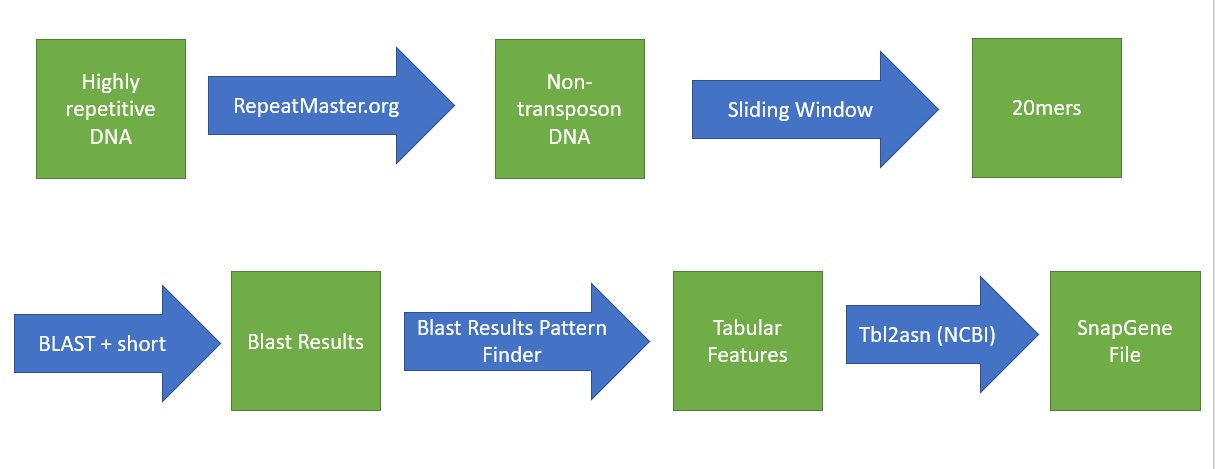
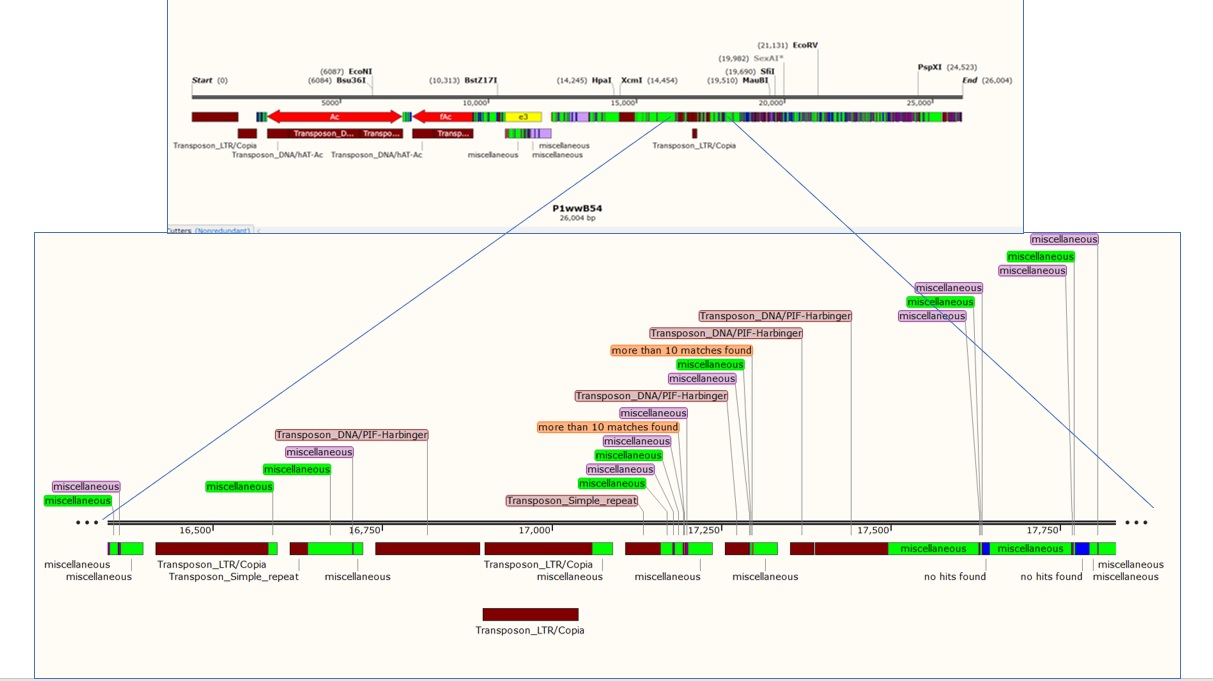
My most recent excited project a research project at Auburn University in Alabama. There I worked with a professor in the Weed and Soil Sciences who has done a lot of work in bioinformatics. In his lab, I did an close analysis of genes in Euphorbia maculata by looking for genes that I knew were in Hevea Brasiliensis, the rubber tree. To accomplish a faster way to do this, I created a BASH script that runs a few existing programs and two python scripts that I created. The program pulls gene data from NCBI, a large biological database, and runs it through a BLAST search to check if it is in the genome of the organism in question. Once the BLAST search identifies a match in a genome, that section of the genome is pulled out and return as a FASTA sequence which can then be analyzed. The python scripts were created using objects from Biopython.
Below is my reserach poster, which showcase more of the biological side of my project.

At Iowa State University, programming classes are typically project based. Because of this, I have created several large java projects that I am very proud of. Some of my favorites include: coding the logic for a game of Snake and a game of Flow, creating a collection of classes of DNA sequence objects for modeling translation, and creating a Football Game class that can simulate a football game.
This website was really fun to create. In my first exposure to HTML, I learned about adding links, photos, and attachments. I learned enough to make a functional website but I am excited to expand my experiences and learn javascript to add more exciting elements to the site.